publications
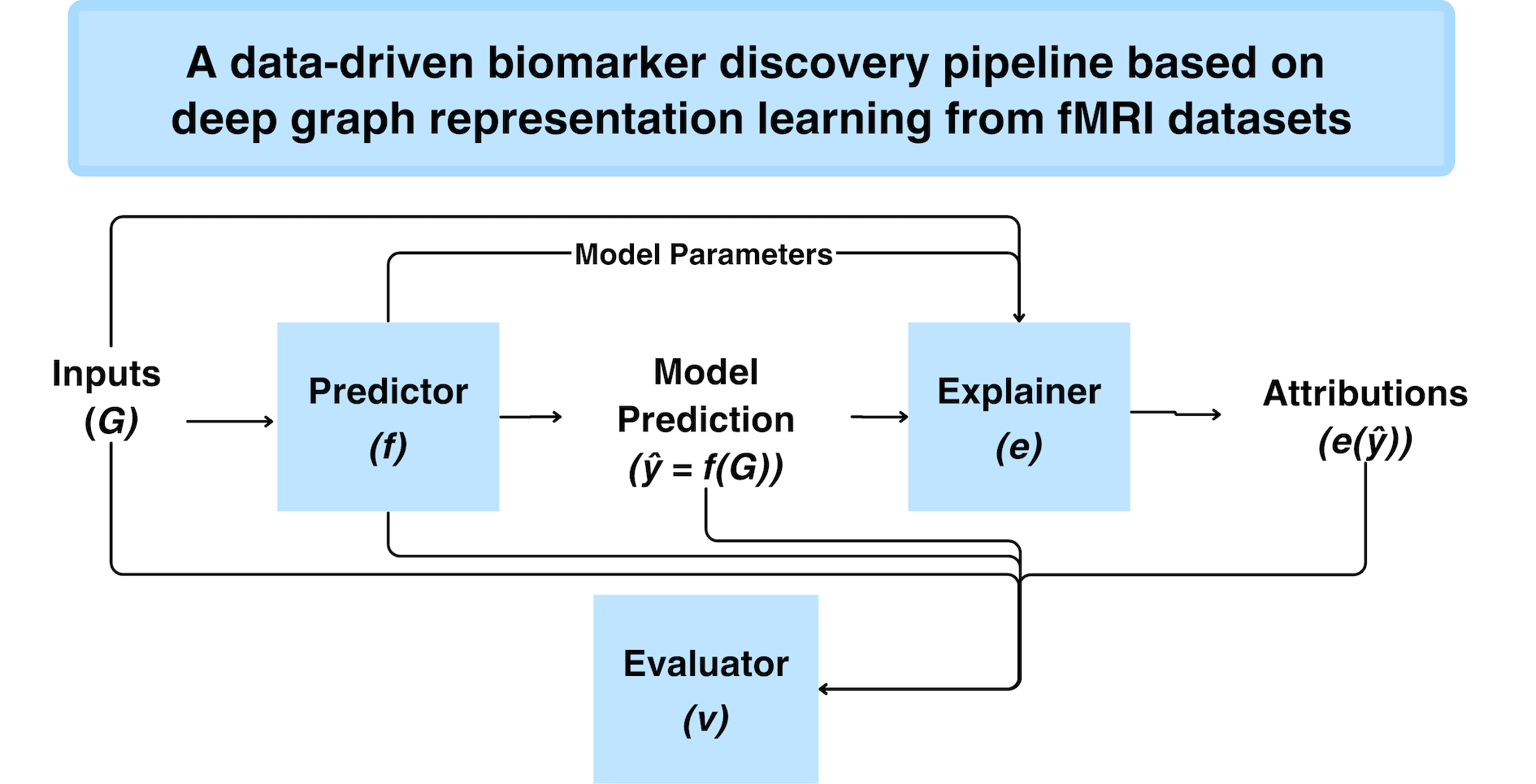
During my PhD studies, I developed techniques to model small and high dimensional multimodal datasets (i.e. large p, small n). Specifically, custom graph neural networks and explainable AI techniques were created to derive connectome-based disease biomarkers (i.e. brain regions/connections affected by diseases). Our techniques go beyond existing class-wide approaches by producing individualised and subgroup-specific insights that would be more clinically relevant than existing solutions. Along the way, we proposed solutions to address problems such as inter-site variability, disease heterogeneity and multimodal fusion.
The next stage of my research is focused on the robustness of these explanations. Most existing studies do not robustly evaluate the salient features highlighted by their algorithms. However, doing so is key towards improving our understanding of neurological disorders. Many obstacles exist, including problems related to data quality (e.g. there’s no agreement on the best fMRI preprocessing pipeline), the perception that neural networks are black boxes and the limited repertoire of tools we have to evaluate explainable AI techniques. We’re actively working on this and if you are keen to collaborate, do send me an email! :)
Beyond connectomes, I have also worked on anatomical brain imaging (i.e. lesion / tissue segmentation) and more pragmatic forms of vision-language models for radiology AI applications. More details will be shared here at a more appropriate time.
2025
- Evaluation
 Discovering robust biomarkers of psychiatric disorders from resting-state functional MRI via graph neural networks: A systematic reviewYi Hao Chan, Deepank Girish, Sukrit Gupta , and 5 more authorsNeuroimage, 2025
Discovering robust biomarkers of psychiatric disorders from resting-state functional MRI via graph neural networks: A systematic reviewYi Hao Chan, Deepank Girish, Sukrit Gupta , and 5 more authorsNeuroimage, 2025Graph neural networks (GNN) have emerged as a popular tool for modeling functional magnetic resonance imaging (fMRI) datasets. Many recent studies have reported significant improvements in disorder classification performance via more sophisticated GNN designs and highlighted salient features that could be potential biomarkers of the disorder. However, existing methods of evaluating their robustness are often limited to cross-referencing with existing literature, which is a subjective and inconsistent process. In this review, we provide an overview of how GNN and model explainability techniques (specifically, feature attributors) have been applied to fMRI datasets for disorder prediction tasks, with an emphasis on evaluating the robustness of potential biomarkers produced by these feature attributors for psychiatric disorders. Then, 65 studies using GNNs that reported potential fMRI biomarkers for psychiatric disorders (attention-deficit hyperactivity disorder, autism spectrum disorder, major depressive disorder, schizophrenia) published before 9 October 2024 were identified from 2 online databases (Scopus, PubMed). We found that while most studies have performant models, salient features highlighted in these studies (as determined by feature attribution scores) vary greatly across studies on the same disorder. Reproducibility of biomarkers is only limited to a small subset at the level of regions and few transdiagnostic biomarkers were identified. To address these issues, we suggest establishing new standards that are based on objective evaluation metrics to determine the robustness of these potential biomarkers. We further highlight gaps in the existing literature and put together a prediction–attribution–evaluation framework that could set the foundations for future research on discovering robust biomarkers of psychiatric disorders via GNNs.
@article{chan2025discovering, title = {Discovering robust biomarkers of psychiatric disorders from resting-state functional MRI via graph neural networks: A systematic review}, author = {Chan, Yi Hao and Girish, Deepank and Gupta, Sukrit and Xia, Jing and Kasi, Chockalingam and He, Yinan and Wang, Conghao and Rajapakse, Jagath C}, journal = {Neuroimage}, year = {2025}, } - Biomarkers
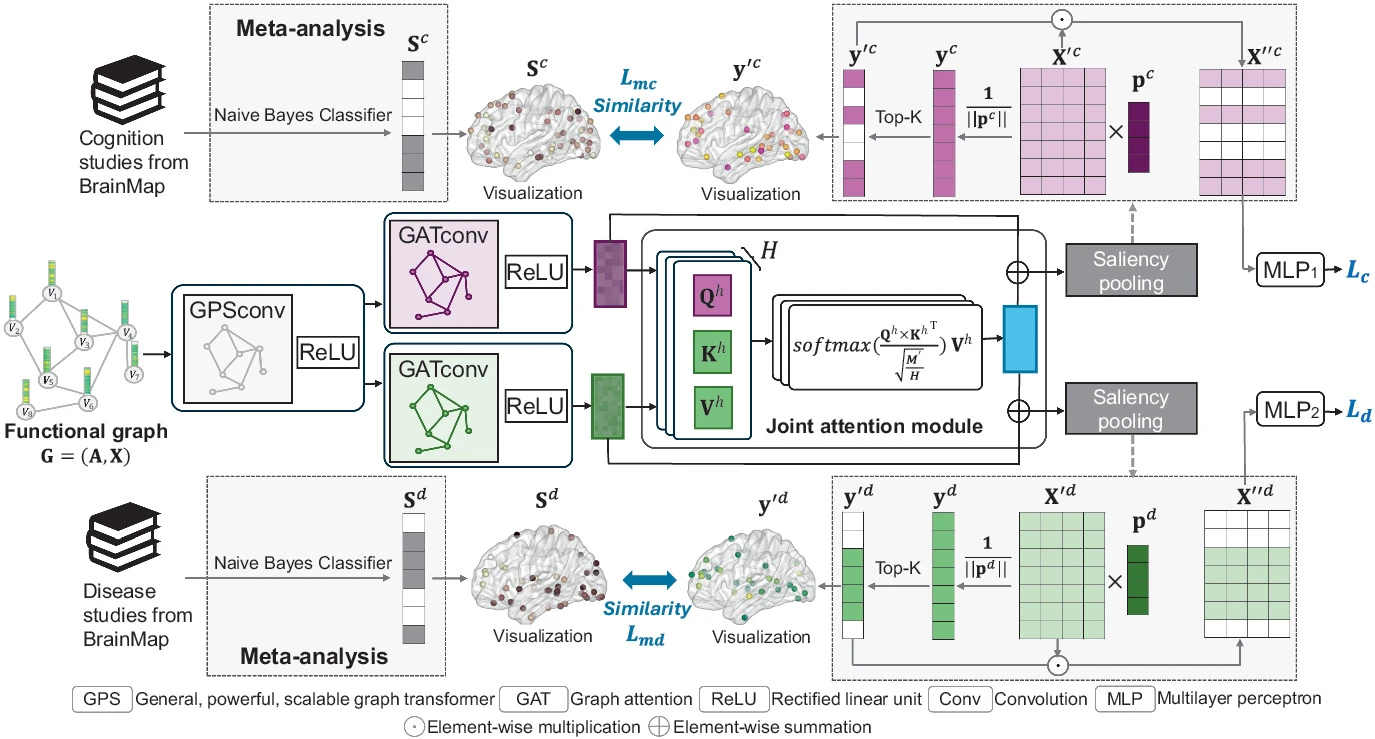 Meta-analysis Guided Multi-task Graph Transformer Network for Diagnosis of Neurological Disease and Cognitive DeficitsJing Xia, Yi Hao Chan, and Jagath C Rajapakse(Highlighted paper) , 2025
Meta-analysis Guided Multi-task Graph Transformer Network for Diagnosis of Neurological Disease and Cognitive DeficitsJing Xia, Yi Hao Chan, and Jagath C Rajapakse(Highlighted paper) , 2025Neurological diseases, such as schizophrenia and attention deficit hyperactivity disorder (ADHD), alter functional connectivity (FC) and are often accompanied by cognitive deficits. Leveraging shared neural mechanisms underlying both neurological disease and cognitive deficits can enhance diagnostic accuracy. However, due to the complex neural mechanisms of these conditions, diagnosing them based on FC alone still presents challenges in terms of accuracy and biomarker reliability. To address these challenges, we designed a meta-analysis guided multi-task graph transformer network to simultaneously predict neurological disease and cognitive deficits and examine alterations in brain FC associated with these conditions. The framework employs a graph transformer method as the encoder and integrates a joint attention mechanism to capture shared disease–cognition features while utilizing saliency pooling to extract saliency weights for each task. To enhance the reliability of saliency weights, we incorporate meta-analysis guidance that aggregates data from 470 functional studies in the BrainMap database. Then, we establish reference probability maps for brain activations associated with neurological diseases and cognitive deficits using a Naive Bayes classifier. The saliency weights learned from saliency pooling are then constrained to align with these references using Pearson correlation. Experiments on the COBRE and ADHD-200 datasets indicate that our proposed method outperforms state-of-the-art multi-task learning models in classifying schizophrenia and ADHD, as well as in predicting their related cognitive deficits. Moreover, the biomarkers extracted from our models exhibit biologically meaningful patterns.
@article{xia2025metaanalysis, title = {Meta-analysis Guided Multi-task Graph Transformer Network for Diagnosis of Neurological Disease and Cognitive Deficits}, author = {Xia, Jing and Chan, Yi Hao and Rajapakse, Jagath C}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, pages = {459--468}, year = {2025}, organization = {Springer}, } - Biomarkers
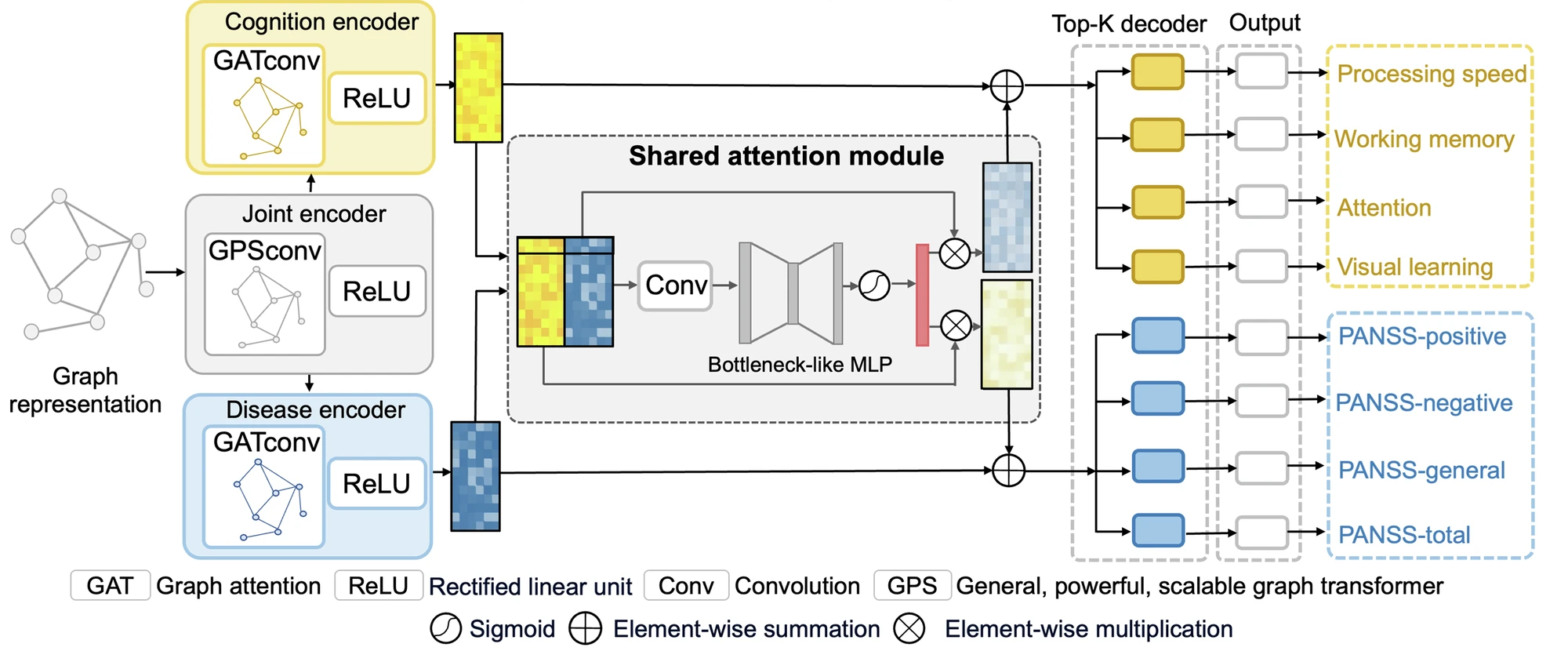 Disentangling shared and unique brain functional changes associated with clinical severity and cognitive phenotypes in schizophrenia via deep learningJing Xia, Yi Hao Chan, Deepank Girish , and 3 more authorsCommunications Biology, 2025
Disentangling shared and unique brain functional changes associated with clinical severity and cognitive phenotypes in schizophrenia via deep learningJing Xia, Yi Hao Chan, Deepank Girish , and 3 more authorsCommunications Biology, 2025Individuals with schizophrenia experience significant cognitive impairments and alterations in brain function. However, the shared and unique brain functional patterns underlying cognition deficits and symptom severity in schizophrenia remain poorly understood. We design an interpretable graph-based multi-task deep learning framework to enhance the simultaneous prediction of schizophrenia illness severity and cognitive functioning measurements by using functional connectivity, and identify both shared and unique brain patterns associated with these phenotypes on 378 subjects from three datasets. Our framework outperforms both single-task and state-of-the-art multi-task learning methods in predicting four Positive and Negative Syndrome Scale (PANSS) subscales and four cognitive domain scores. The performance is replicable across three datasets, and the shared and unique functional changes are confirmed by meta-analysis at both regional and modular levels. Our study provides insights into the neural correlates of illness severity and cognitive implications, offering potential targets for further evaluations of treatment effects and longitudinal follow-up.
@article{xia2025disentangling, title = {Disentangling shared and unique brain functional changes associated with clinical severity and cognitive phenotypes in schizophrenia via deep learning}, author = {Xia, Jing and Chan, Yi Hao and Girish, Deepank and Chew, Qian Hui and Sim, Kang and Rajapakse, Jagath C}, journal = {Communications Biology}, volume = {8}, number = {1}, pages = {1215}, year = {2025}, publisher = {Nature Publishing Group UK London}, } - Multimodal
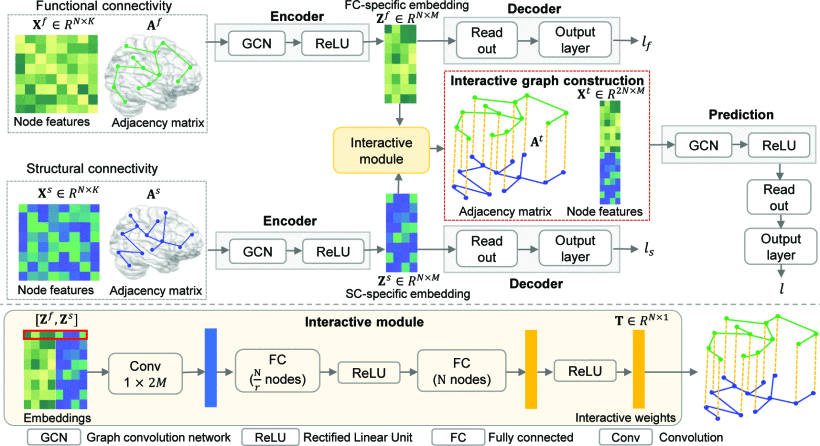
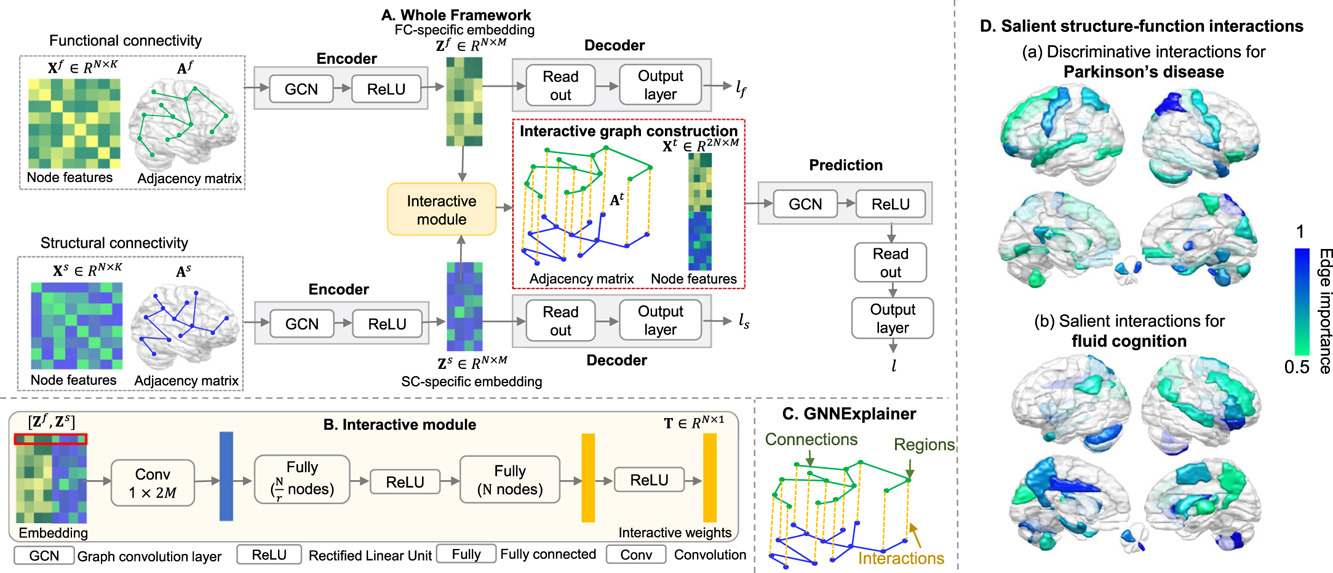 Interpretable modality-specific and interactive graph convolutional network on brain functional and structural connectomesJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorMedical Image Analysis, 2025
Interpretable modality-specific and interactive graph convolutional network on brain functional and structural connectomesJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorMedical Image Analysis, 2025Both brain functional connectivity (FC) and structural connectivity (SC) provide distinct neural mechanisms for cognition and neurological disease. In addition, interactions between SC and FC within distributed association regions are related to alterations in cognition or neurological diseases, considering the inherent linkage between neural function and structure. However, there is a scarcity of existing learning-based methods that leverage both modality-specific characteristics and high-order interactions between the two modalities for regression or classification. Hence, this study proposes an interpretable modality-specific and interactive graph convolutional network (MS-Inter-GCN) that incorporates modality-specific information, reflecting the unique neural mechanism for each modality, and structure–function interactions, capturing the underlying foundation provided by white-matter fiber tracts for high-level brain function. In MS-Inter-GCN, we generate modality-specific task-relevant embeddings separately from both FC and SC using a graph convolutional encoder–decoder module. Subsequently, we learn the interactive weights between corresponding regions of FC and SC, reflecting the coupling strength, by employing an interactive module on the embeddings of both modalities. A novel graph structure is constructed, which uses modality-specific task-relevant embeddings and inserts the interactive weights as edges connecting corresponding regions of two modalities, and then is used for the regression or classification task. Finally, a post-hoc explainable technology - GNNExplainer- is used to identify salient regions and connections of each modality as well as salient interactions between FC and SC associated with tasks. We apply the proposed framework to fluid cognition prediction, Parkinson’s disease (PD), Alzheimer’s disease (AD), and schizophrenia (SZ) classification. Experimental results demonstrate that our method outperforms the other ten state-of-the-art methods on multi-modal brain features on all tasks. The GNNExplainer identifies salient structural and functional regions and connections for fluid cognition, PD, AD, and SZ. It confirms that strong structure–function coupling within the executive and control networks, combined with weak coupling within the motor network, is associated with fluid cognition. Moreover, structure–function decoupling in specific brain regions serves as a marker for different diseases: decoupling of the prefrontal, superior parietal, and superior occipital cortices is a marker of PD; decoupling of the middle frontal and lateral parietal cortices, temporal pole, and subcortical regions is indicative of AD; and decoupling of the prefrontal, parietal, and temporal cortices, as well as the cerebellum, contributes to SZ.
@article{xia2025interpretable, title = {Interpretable modality-specific and interactive graph convolutional network on brain functional and structural connectomes}, author = {Xia, Jing and Chan, Yi Hao and Girish, Deepank and Rajapakse, Jagath C}, journal = {Medical Image Analysis}, volume = {102}, pages = {103509}, year = {2025}, publisher = {Elsevier}, } - Multimodal
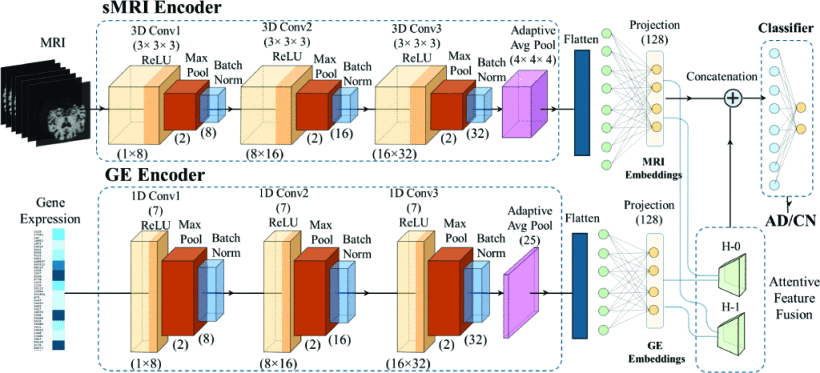 Decoding Brain Structure and Gene Expression Interactions in Alzheimer’s Disease PathologyAmashi Niwarthana, Chockalingam Kasi, Yi Hao Chan , and 2 more authorsIn ICASSP 2025-2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) , 2025
Decoding Brain Structure and Gene Expression Interactions in Alzheimer’s Disease PathologyAmashi Niwarthana, Chockalingam Kasi, Yi Hao Chan , and 2 more authorsIn ICASSP 2025-2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) , 2025Although brain imaging has provided crucial insights into Alzheimer’s disease (AD) progression, its limitations in capturing molecular changes have motivated the need to integrate and interpret transcriptomic data, which can reveal early-stage molecular disruptions. By interpreting both structural and gene expression patterns, there is potential to gain a more comprehensive understanding of the disease and identify interactions that drive AD pathogenesis. This study introduces a novel deep learning framework that integrates structural magnetic resonance imaging (sMRI) and gene expression (GE) data for AD prediction. The model extracts features from both data modalities and employs a novel approach to quantify cross-modal interactions. Our multimodal architecture achieves 83.6% accuracy in AD prediction, demonstrating its effectiveness in combining imaging and transcriptomic data. We present a technique for interpreting interactions between specific brain regions and gene expression patterns, providing insights into their joint contribution to AD risk. This approach enables the identification of both established and potential new AD biomarkers, spanning structural brain changes and transcriptomic alterations. By offering a deeper understanding of AD’s molecular and structural aspects, our work advances the field of multimodal biomarker discovery in neurodegenerative diseases and paves the way for more comprehensive early diagnosis strategies.
@inproceedings{niwarthana2025decoding, title = {Decoding Brain Structure and Gene Expression Interactions in Alzheimer’s Disease Pathology}, author = {Niwarthana, Amashi and Kasi, Chockalingam and Chan, Yi Hao and Wang, Conghao and Rajapakse, Jagath C}, booktitle = {ICASSP 2025-2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)}, pages = {1--5}, year = {2025}, organization = {IEEE}, }
2024
- Evaluation
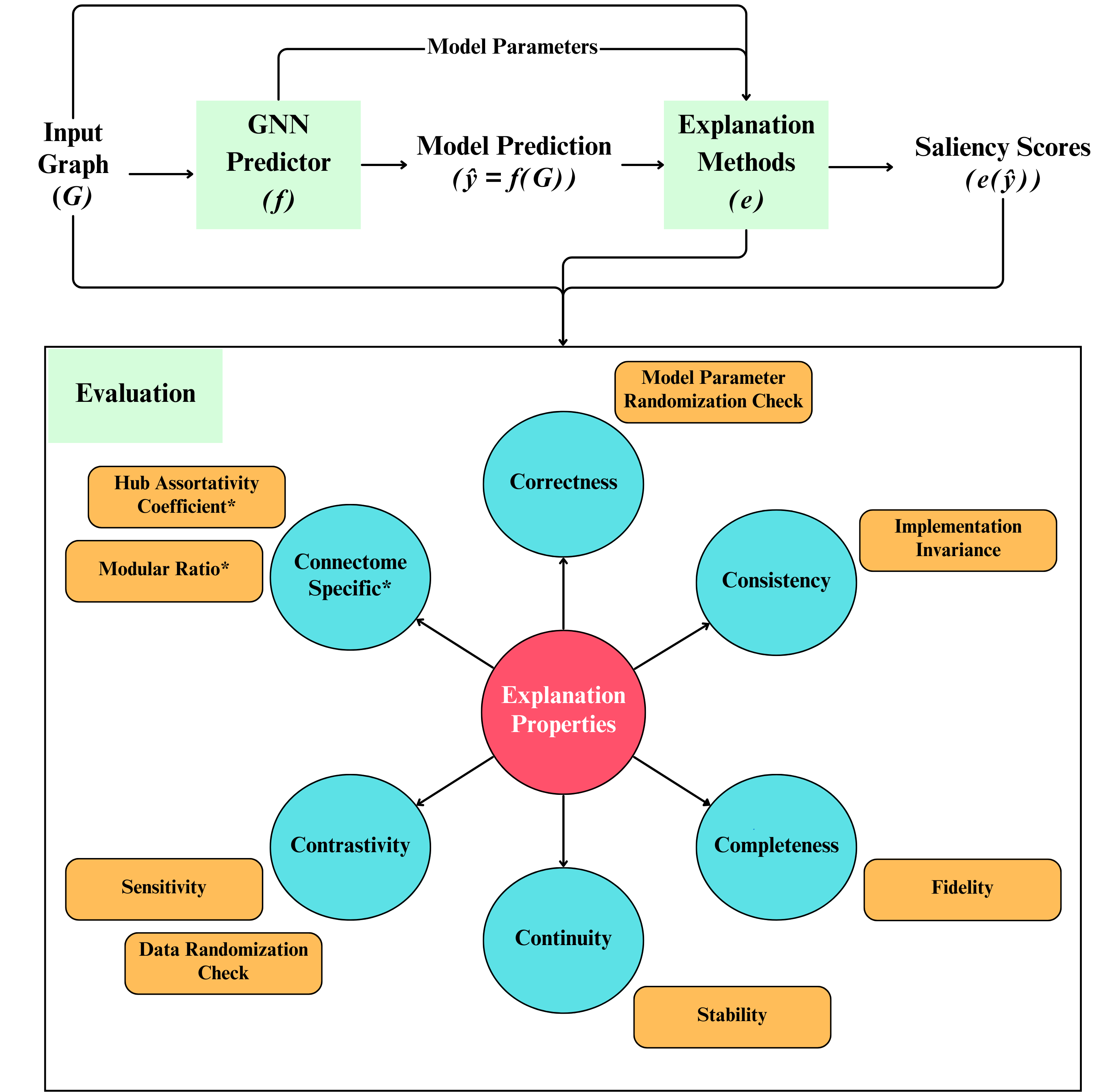 Robustness of explainable AI algorithms for disease biomarker discovery from functional connectivity datasetsDeepank Girish, Yi Hao Chan, Sukrit Gupta , and 2 more authorsIn 2024 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI) , 2024
Robustness of explainable AI algorithms for disease biomarker discovery from functional connectivity datasetsDeepank Girish, Yi Hao Chan, Sukrit Gupta , and 2 more authorsIn 2024 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI) , 2024Machine learning (ML) models have been used in functional neuroimaging for wide-ranging tasks, ranging from disease diagnosis to disease prognosis. There have been successive functional connectivity-based ML studies focused on improving model performances for disease detection. An increasing number of such studies use their trained models to detect and evaluate salient features that could be potential biomarkers of these neuro-logical conditions. The evaluation of these salient features is often qualitative and limited to cross-referencing existing literature for similar findings. In this study, we present objective quantitative metrics to evaluate the robustness of these salient features. Building upon existing generic evaluation metrics, we propose metrics that capture topological properties known to be characteristic of brain functional connectomes. Using existing and newly proposed measures on a set of baselines and state-of-the-art graph neural networks (GNN) models, we found that when GNNExplainer is used with models that incorporate attention, the scores produced are relatively more robust than other combinations. On datasets of patients with Autism Spectrum Disorder (ASD) or Attention-deficit Hyperactivity Disorder (ADHD), our proposed metrics highlighted that salient features identified in both disorders are highly involved in functional specialization, while salient ASD features expressed stronger functional integration than ADHD. We package these existing and novel metrics together in the RE-CONFIRM framework that holds promise to set the foundations for the quantitative evaluation of salient features detected by future studies.
@inproceedings{girish2024robustness, title = {Robustness of explainable AI algorithms for disease biomarker discovery from functional connectivity datasets}, author = {Girish, Deepank and Chan, Yi Hao and Gupta, Sukrit and Xia, Jing and Rajapakse, Jagath C}, booktitle = {2024 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI)}, pages = {1--8}, year = {2024}, organization = {IEEE}, } - Multimodal
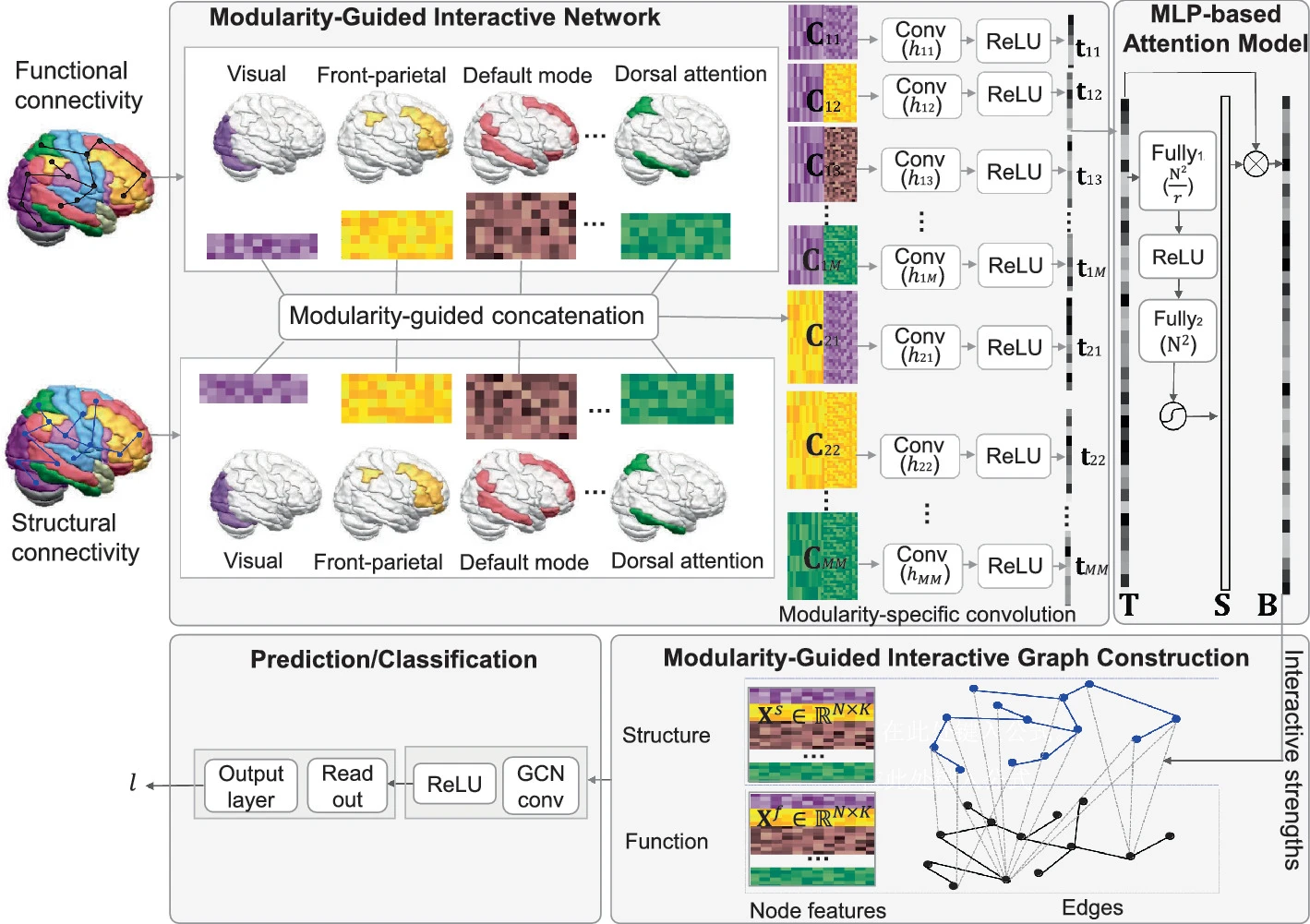 IMG-GCN: Interpretable modularity-guided structure-function interactions learning for brain cognition and disorder analysisJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorIn International Conference on Medical Image Computing and Computer-Assisted Intervention (Early accept) , 2024
IMG-GCN: Interpretable modularity-guided structure-function interactions learning for brain cognition and disorder analysisJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorIn International Conference on Medical Image Computing and Computer-Assisted Intervention (Early accept) , 2024Brain structure-function interaction is crucial for cognition and brain disorder analysis, and it is inherently more complex than a simple region-to-region coupling. It exhibits homogeneity at the modular level, with regions of interest (ROIs) within the same module showing more similar neural mechanisms than those across modules. Leveraging modular-level guidance to capture complex structure-function interactions is essential, but such studies are still scarce. Therefore, we propose an interpretable modularity-guided graph convolution network (IMG-GCN) to extract the structure-function interactions across ROIs and highlight the most discriminative interactions relevant to fluid cognition and Parkinson’s disease (PD). Specifically, we design a modularity-guided interactive network that defines modularity-specific convolution operation to learn interactions between structural and functional ROIs according to modular homogeneity. Then, an MLP-based attention model is introduced to identify the most contributed interactions. The interactions are inserted as edges linking structural and functional ROIs to construct a unified combined graph, and GCN is applied for final tasks. Experiments on HCP and PPMI datasets indicate that our proposed method outperforms state-of-the-art multi-model methods in fluid cognition prediction and PD classification. The attention maps reveal that the frontoparietal and default mode structures interacting with visual function are discriminative for fluid cognition, while the subcortical structures interacting with widespread functional modules are associated with PD.
@inproceedings{xia2024img, title = {IMG-GCN: Interpretable modularity-guided structure-function interactions learning for brain cognition and disorder analysis}, author = {Xia, Jing and Chan, Yi Hao and Girish, Deepank and Rajapakse, Jagath C}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, pages = {470--480}, year = {2024}, organization = {Springer}, } - Segmentation
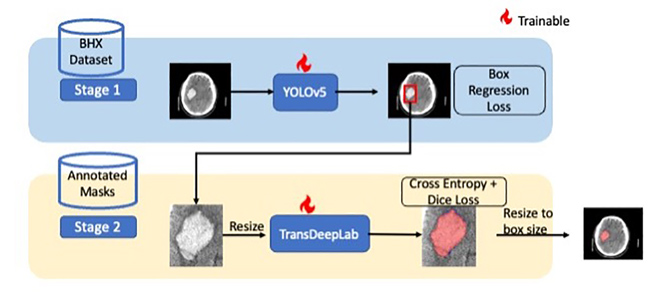 Two-stage approach to intracranial hemorrhage segmentation from head CT imagesJagath C Rajapakse, Chun Hung How, Yi Hao Chan , and 5 more authorsIEEE Access, 2024
Two-stage approach to intracranial hemorrhage segmentation from head CT imagesJagath C Rajapakse, Chun Hung How, Yi Hao Chan , and 5 more authorsIEEE Access, 2024Intracranial hemorrhage (ICH) is an emergency and a potentially life-threatening condition. Automated segmentation of ICH from head CT images can provide clinicians with volumetric measures that can be used for diagnosis and decision support for treatment procedures. Existing solutions typically involve training deep learning models to perform segmentation directly on the whole CT image. However, datasets with segmentation masks are typically very small in comparison with datasets with bounding boxes. Thus, we propose a two-stage approach that utilizes both bounding boxes and segmentation masks to help improve segmentation performance. In the first stage, ICH regions are detected and localized with bounding boxes surrounding the lesion by using a supervised YOLOv5 object detector. In the second stage, the localized ICH foreground is automatically segmented using TransDeepLab, an attention-based transformer network. Although we utilize both ground-truth bounding boxes and segmentation masks, different datasets can be used to train each stage. There is no requirement for pairing up bounding boxes and segmentation masks to train the model. Since bounding box annotations are available in larger quantities than segmentation masks, our approach allows these large datasets of bounding boxes to be used to improve ICH segmentation performance. On our dataset of segmentation masks, we demonstrated that our proposed two-stage YOLOv5+ TransDeepLab model outperformed segmentation methods such as SegResNet by 8% in terms of Dice score. Given ground truth bounding boxes, a Dice score of 0.769 is achieved, outperforming state-of-the-art methods such as nnU-Net. In sum, our proposed two-stage approach produces more accurate binary segmentation of ICH for neuroradiologists and these improved measurements could potentially aid their clinical decision-making process.
@article{rajapakse2024two, title = {Two-stage approach to intracranial hemorrhage segmentation from head CT images}, author = {Rajapakse, Jagath C and How, Chun Hung and Chan, Yi Hao and Hao, Luke Chin Peng and Padhi, Abhinandan and Adrakatti, Vivek and Khan, Iram and Lim, Tchoyoson}, journal = {IEEE Access}, year = {2024}, publisher = {IEEE}, } - Multimodal
 Brain Structure-Function Interaction Network for Fluid Cognition PredictionJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorIn ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Oral) , 2024
Brain Structure-Function Interaction Network for Fluid Cognition PredictionJing Xia, Yi Hao Chan, Deepank Girish , and 1 more authorIn ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Oral) , 2024Predicting fluid cognition via neuroimaging data is essential for understanding the neural mechanisms underlying various complex cognitions in the human brain. Both brain functional connectivity (FC) and structural connectivity (SC) provide distinct neural mechanisms for fluid cognition. In addition, interactions between SC and FC within distributed association regions are related to improvements in fluid cognition. However, existing learning-based methods that leverage both modality-specific embeddings and high-order interactions between the two modalities for prediction are scarce. To tackle these challenges, this study proposes an end-to-end brain structure-function interaction network that incorporates both modality-specific embeddings and structure-function interactions to predict fluid cognition. In this model, we generate embeddings from both FC and SC separately using a graph convolution encoder-decoder module. Subsequently, we learn the interactive weights between corresponding regions of FC and SC, reflecting the coupling strength, by employing an interactive module on the embeddings of both modalities. A novel graph structure - utilizing modality-specific embeddings and interactive weights - is constructed and used for the final prediction. Experimental results demonstrate that our proposed method outperforms other state-of-the-art methods employed on uni-modal and multi-modal brain features. We further identify that strong structure-function coupling in the inferior frontal, postcentral, superior temporal and cingulate cortices are associated with fluid intelligence.
@inproceedings{xia2024brain, title = {Brain Structure-Function Interaction Network for Fluid Cognition Prediction}, author = {Xia, Jing and Chan, Yi Hao and Girish, Deepank and Rajapakse, Jagath C}, booktitle = {ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)}, pages = {1706--1710}, year = {2024}, organization = {IEEE}, } - Biomarkers
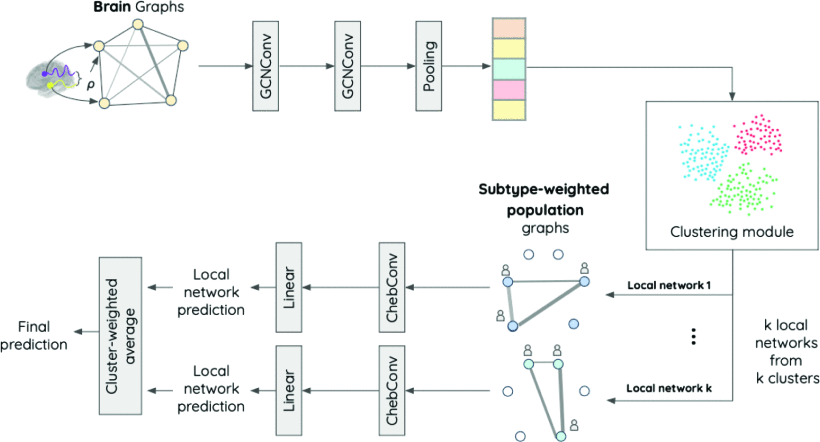 Subtype-Specific Biomarkers of Alzheimer’s Disease from Anatomical and Functional Connectomes via Graph Neural NetworksYi Hao Chan, Jun Liang Ang, Sukrit Gupta , and 2 more authorsIn ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Oral) , 2024
Subtype-Specific Biomarkers of Alzheimer’s Disease from Anatomical and Functional Connectomes via Graph Neural NetworksYi Hao Chan, Jun Liang Ang, Sukrit Gupta , and 2 more authorsIn ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Oral) , 2024Heterogeneity is present in Alzheimer’s disease (AD), making it challenging to study. To address this, we propose a graph neural network (GNN) approach to identify disease subtypes from magnetic resonance imaging (MRI) and functional MRI (fMRI) scans. Subtypes are identified by encoding the patients’ scans in brain graphs (via cortical similarity networks) and clustering the representations learnt by the GNN. These subtyping information are used to construct population graphs for an ensemble of local networks, each producing intermediate predictions that are subsequently combined to produce the model’s final decision. Using MRI and fMRI scans from two datasets on AD, we demonstrate that our proposed architecture outperforms existing methods. Three subtypes of AD were identified and left cuneus was found to be a consistent class-wide biomarker. Subtype-specific biomarkers produced by our method further revealed deeper insights, including a unique subtype with significant degeneration in the left isthmus cingulate cortex.
@inproceedings{chan2024subtype, title = {Subtype-Specific Biomarkers of Alzheimer’s Disease from Anatomical and Functional Connectomes via Graph Neural Networks}, author = {Chan, Yi Hao and Ang, Jun Liang and Gupta, Sukrit and He, Yinan and Rajapakse, Jagath C}, booktitle = {ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)}, pages = {2195--2199}, year = {2024}, organization = {IEEE}, }
2023
- Biomarkers
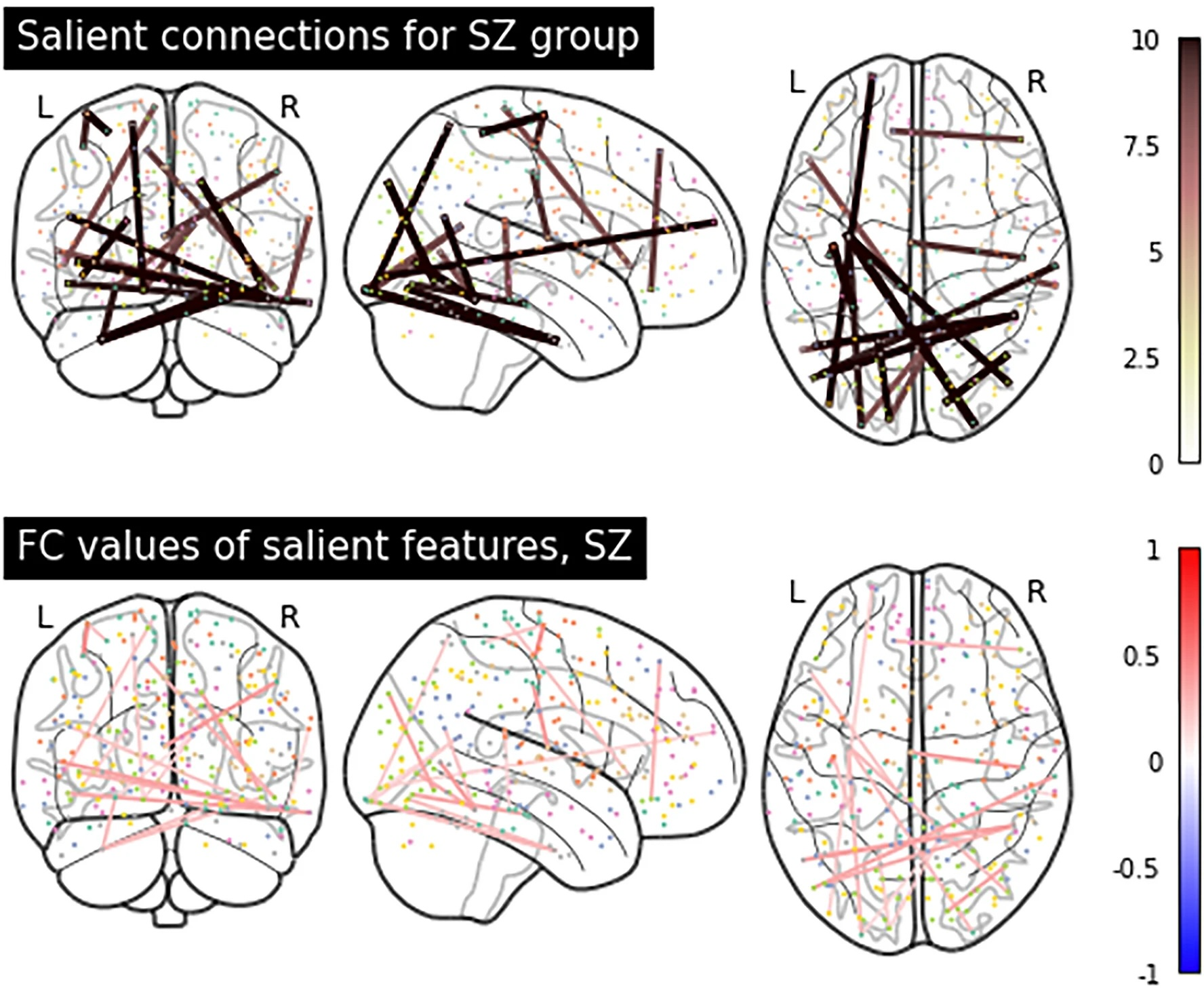 Elucidating salient site-specific functional connectivity features and site-invariant biomarkers in schizophrenia via deep neural networksYi Hao Chan, Wei Chee Yew, Qian Hui Chew , and 2 more authorsScientific Reports, 2023
Elucidating salient site-specific functional connectivity features and site-invariant biomarkers in schizophrenia via deep neural networksYi Hao Chan, Wei Chee Yew, Qian Hui Chew , and 2 more authorsScientific Reports, 2023Schizophrenia is a highly heterogeneous disorder and salient functional connectivity (FC) features have been observed to vary across study sites, warranting the need for methods that can differentiate between site-invariant FC biomarkers and site-specific salient FC features. We propose a technique named Semi-supervised learning with data HaRmonisation via Encoder-Decoder-classifier (SHRED) to examine these features from resting state functional magnetic resonance imaging scans gathered from four sites. Our approach involves an encoder-decoder-classifier architecture that simultaneously performs data harmonisation and semi-supervised learning (SSL) to deal with site differences and labelling inconsistencies across sites respectively. The minimisation of reconstruction loss from SSL was shown to improve model performance even within small datasets whilst data harmonisation often led to lower model generalisability, which was unaffected using the SHRED technique. We show that our proposed model produces site-invariant biomarkers, most notably the connection between transverse temporal gyrus and paracentral lobule. Site-specific salient FC features were also elucidated, especially implicating the paracentral lobule for our local dataset. Our examination of these salient FC features demonstrates how site-specific features and site-invariant biomarkers can be differentiated, which can deepen our understanding of the neurobiology of schizophrenia.
@article{chan2023elucidating, title = {Elucidating salient site-specific functional connectivity features and site-invariant biomarkers in schizophrenia via deep neural networks}, author = {Chan, Yi Hao and Yew, Wei Chee and Chew, Qian Hui and Sim, Kang and Rajapakse, Jagath C}, journal = {Scientific Reports}, volume = {13}, number = {1}, pages = {21047}, year = {2023}, publisher = {Nature Publishing Group UK London}, } - Multimodal
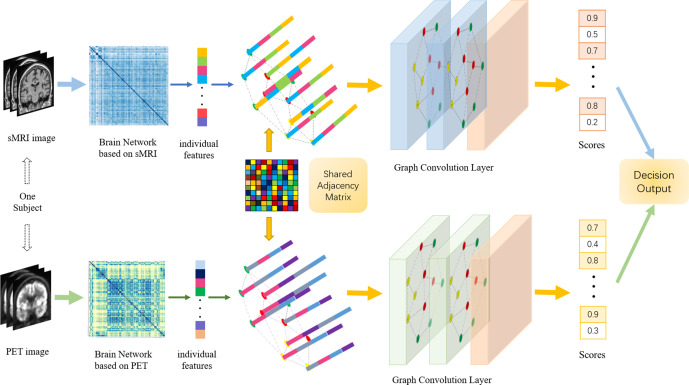 Multi-modal graph neural network for early diagnosis of Alzheimer’s disease from sMRI and PET scansYanteng Zhang, Xiaohai He, Yi Hao Chan , and 2 more authorsComputers in Biology and Medicine, 2023
Multi-modal graph neural network for early diagnosis of Alzheimer’s disease from sMRI and PET scansYanteng Zhang, Xiaohai He, Yi Hao Chan , and 2 more authorsComputers in Biology and Medicine, 2023In recent years, deep learning models have been applied to neuroimaging data for early diagnosis of Alzheimer’s disease (AD). Structural magnetic resonance imaging (sMRI) and positron emission tomography (PET) images provide structural and functional information about the brain, respectively. Combining these features leads to improved performance than using a single modality alone in building predictive models for AD diagnosis. However, current multi-modal approaches in deep learning, based on sMRI and PET, are mostly limited to convolutional neural networks, which do not facilitate integration of both image and phenotypic information of subjects. We propose to use graph neural networks (GNN) that are designed to deal with problems in non-Euclidean domains. In this study, we demonstrate how brain networks are created from sMRI or PET images and can be used in a population graph framework that combines phenotypic information with imaging features of the brain networks. Then, we present a multi-modal GNN framework where each modality has its own branch of GNN and a technique that combines the multi-modal data at both the level of node vectors and adjacency matrices. Finally, we perform late fusion to combine the preliminary decisions made in each branch and produce a final prediction. As multi-modality data becomes available, multi-source and multi-modal is the trend of AD diagnosis. We conducted explorative experiments based on multi-modal imaging data combined with non-imaging phenotypic information for AD diagnosis and analyzed the impact of phenotypic information on diagnostic performance. Results from experiments demonstrated that our proposed multi-modal approach improves performance for AD diagnosis. Our study also provides technical reference and support the need for multivariate multi-modal diagnosis methods.
@article{zhang2023multi, title = {Multi-modal graph neural network for early diagnosis of Alzheimer's disease from sMRI and PET scans}, author = {Zhang, Yanteng and He, Xiaohai and Chan, Yi Hao and Teng, Qizhi and Rajapakse, Jagath C}, journal = {Computers in Biology and Medicine}, volume = {164}, pages = {107328}, year = {2023}, publisher = {Elsevier}, }
2022
- Biomarkers
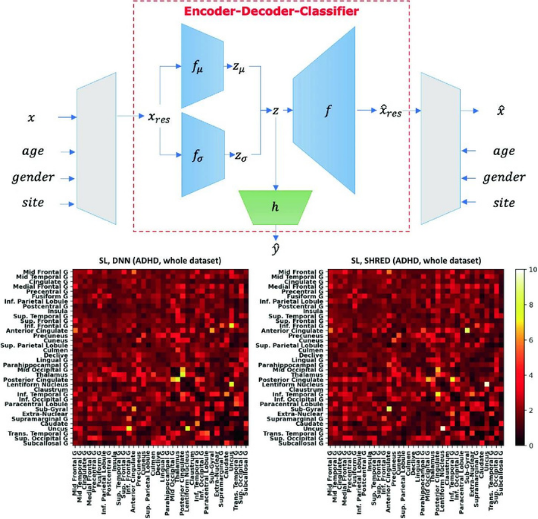 Semi-supervised learning with data harmonisation for biomarker discovery from resting state fMRIYi Hao Chan, Wei Chee Yew, and Jagath C RajapakseIn International Conference on Medical Image Computing and Computer-Assisted Intervention , 2022
Semi-supervised learning with data harmonisation for biomarker discovery from resting state fMRIYi Hao Chan, Wei Chee Yew, and Jagath C RajapakseIn International Conference on Medical Image Computing and Computer-Assisted Intervention , 2022Computational models often overfit on neuroimaging datasets (which are high-dimensional and consist of small sample sizes), resulting in poor inferences such as ungeneralisable biomarkers. One solution is to pool datasets (of similar disorders) from other sites to augment the small dataset, but such efforts have to handle variations introduced by site effects and inconsistent labelling. To overcome these issues, we propose an encoder-decoder-classifier architecture that combines semi-supervised learning with harmonisation of data across sites. The architecture is trained end-to-end via a novel multi-objective loss function. Using the architecture on multi-site fMRI datasets such as ADHD-200 and ABIDE, we obtained significant improvement on classification performance and showed how site-invariant biomarkers were disambiguated from site-specific ones. Our findings demonstrate the importance of accounting for both site effects and labelling inconsistencies when combining datasets from multiple sites to overcome the paucity of data. With the proliferation of neuroimaging research conducted on retrospectively aggregated datasets, our architecture offers a solution to handle site differences and labelling inconsistencies in such datasets. Code is available at https://github.com/SCSE-Biomedical-Computing-Group/SHRED.
@inproceedings{chan2022semi, title = {Semi-supervised learning with data harmonisation for biomarker discovery from resting state fMRI}, author = {Chan, Yi Hao and Yew, Wei Chee and Rajapakse, Jagath C}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, pages = {441--451}, year = {2022}, organization = {Springer}, } - Multimodal
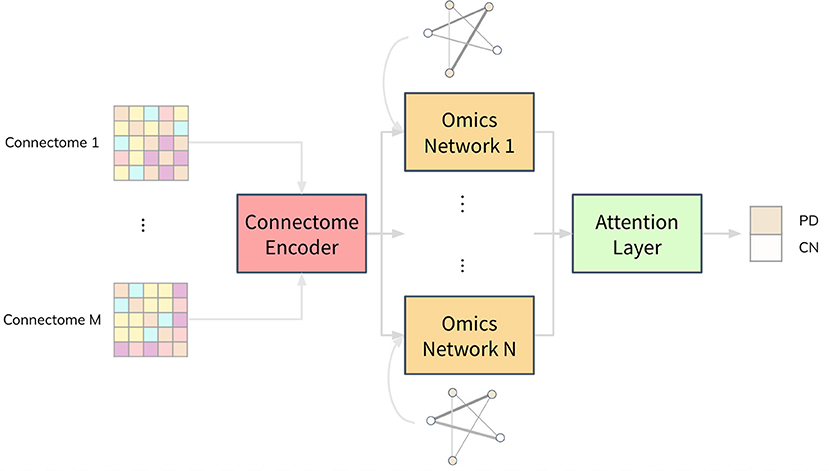 Combining neuroimaging and omics datasets for disease classification using graph neural networksYi Hao Chan, Conghao Wang, Wei Kwek Soh , and 1 more authorFrontiers in Neuroscience, 2022
Combining neuroimaging and omics datasets for disease classification using graph neural networksYi Hao Chan, Conghao Wang, Wei Kwek Soh , and 1 more authorFrontiers in Neuroscience, 2022Both neuroimaging and genomics datasets are often gathered for the detection of neurodegenerative diseases. Huge dimensionalities of neuroimaging data as well as omics data pose tremendous challenge for methods integrating multiple modalities. There are few existing solutions that can combine both multi-modal imaging and multi-omics datasets to derive neurological insights. We propose a deep neural network architecture that combines both structural and functional connectome data with multi-omics data for disease classification. A graph convolution layer is used to model functional magnetic resonance imaging (fMRI) and diffusion tensor imaging (DTI) data simultaneously to learn compact representations of the connectome. A separate set of graph convolution layers are then used to model multi-omics datasets, expressed in the form of population graphs, and combine them with latent representations of the connectome. An attention mechanism is used to fuse these outputs and provide insights on which omics data contributed most to the model’s classification decision. We demonstrate our methods for Parkinson’s disease (PD) classification by using datasets from the Parkinson’s Progression Markers Initiative (PPMI). PD has been shown to be associated with changes in the human connectome and it is also known to be influenced by genetic factors. We combine DTI and fMRI data with multi-omics data from RNA Expression, Single Nucleotide Polymorphism (SNP), DNA Methylation and non-coding RNA experiments. A Matthew Correlation Coefficient of greater than 0.8 over many combinations of multi-modal imaging data and multi-omics data was achieved with our proposed architecture. To address the paucity of paired multi-modal imaging data and the problem of imbalanced data in the PPMI dataset, we compared the use of oversampling against using CycleGAN on structural and functional connectomes to generate missing imaging modalities. Furthermore, we performed ablation studies that offer insights into the importance of each imaging and omics modality for the prediction of PD. Analysis of the generated attention matrices revealed that DNA Methylation and SNP data were the most important omics modalities out of all the omics datasets considered. Our work motivates further research into imaging genetics and the creation of more multi-modal imaging and multi-omics datasets to study PD and other complex neurodegenerative diseases.
@article{chan2022combining, title = {Combining neuroimaging and omics datasets for disease classification using graph neural networks}, author = {Chan, Yi Hao and Wang, Conghao and Soh, Wei Kwek and Rajapakse, Jagath C}, journal = {Frontiers in Neuroscience}, volume = {16}, pages = {866666}, year = {2022}, publisher = {Frontiers Media SA}, }
2021
- Biomarkers
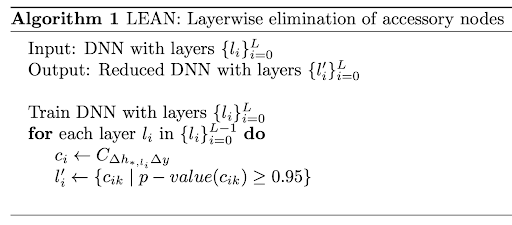 Obtaining leaner deep neural networks for decoding brain functional connectome in a single shotSukrit Gupta*, Yi Hao Chan*, Jagath C Rajapakse , and 2 more authorsNeurocomputing, 2021
Obtaining leaner deep neural networks for decoding brain functional connectome in a single shotSukrit Gupta*, Yi Hao Chan*, Jagath C Rajapakse , and 2 more authorsNeurocomputing, 2021Neuroscientific knowledge points to the presence of redundancy in the correlations of the brain’s functional activity. These redundancies can be removed to mitigate the problem of overfitting when deep neural network (DNN) models are used to classify neuroimaging datasets. We propose an algorithm that removes insignificant nodes of DNNs in a layerwise manner and then adds a subset of correlated features in a single shot. When performing experiments with functional MRI datasets for classifying patients from healthy controls, we were able to obtain simpler and more generalizable DNNs. The obtained DNNs maintained a similar performance as the full network with only around 2% of the initial trainable parameters. Further, we used the trained network to identify salient brain regions and connections from functional connectome for multiple brain disorders. The identified biomarkers were found to closely correspond to previously known disease biomarkers. The proposed methods have cross-modal applications in obtaining leaner DNNs that seem to fit neuroimaging data better. The corresponding code is available at https://github.com/SCSE-Biomedical-Computing-Group/LEAN_CLIP.
@article{gupta2021obtaining, title = {Obtaining leaner deep neural networks for decoding brain functional connectome in a single shot}, author = {Gupta, Sukrit and Chan, Yi Hao and Rajapakse, Jagath C and Initiative, Alzheimer’s Disease Neuroimaging and others}, journal = {Neurocomputing}, volume = {453}, pages = {326--336}, year = {2021}, publisher = {Elsevier}, }
2020
- Time Series
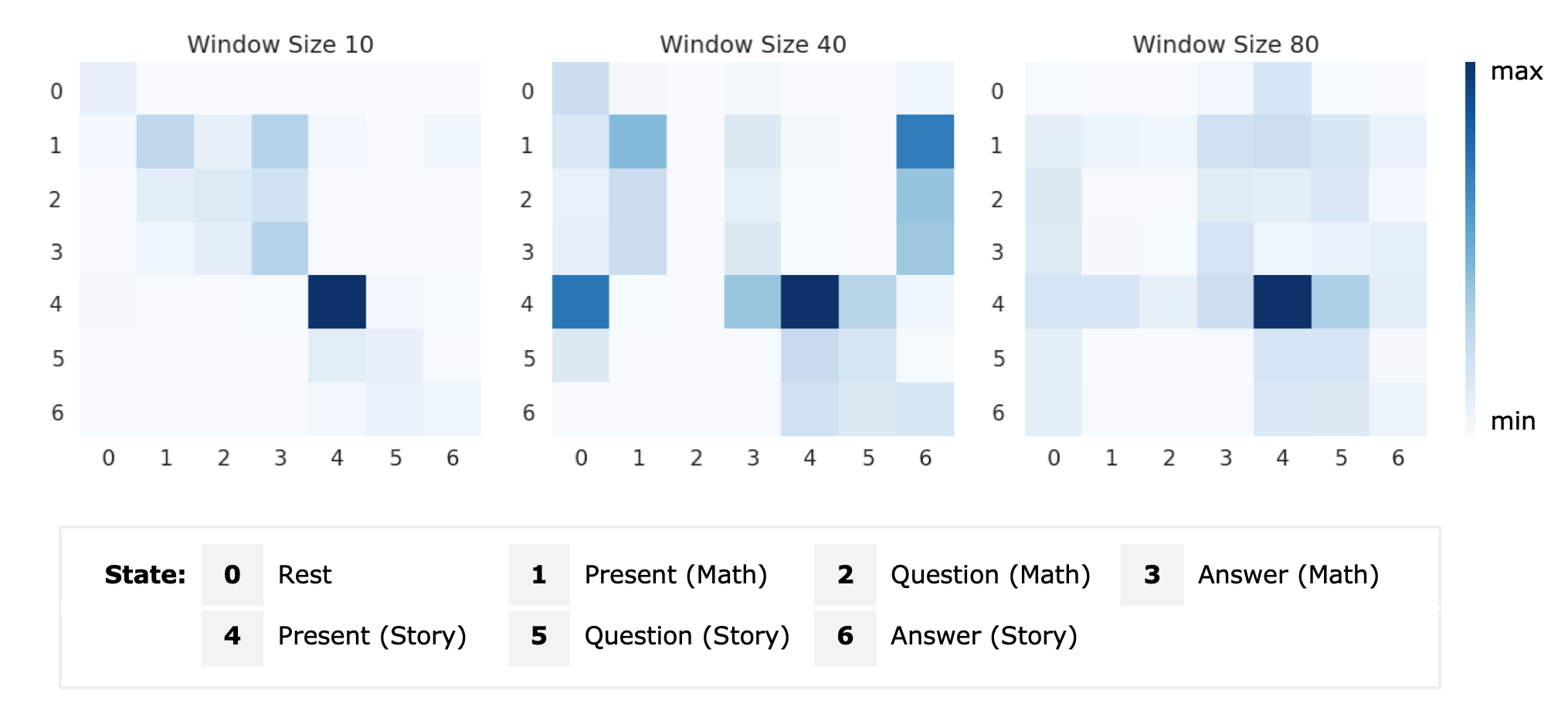 Decoding task states by spotting salient patterns at time points and brain regionsYi Hao Chan, Sukrit Gupta, LL Chamara Kasun , and 1 more authorIn Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology: Third International Workshop, MLCN 2020, and Second International Workshop, RNO-AI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings 3 , 2020
Decoding task states by spotting salient patterns at time points and brain regionsYi Hao Chan, Sukrit Gupta, LL Chamara Kasun , and 1 more authorIn Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology: Third International Workshop, MLCN 2020, and Second International Workshop, RNO-AI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings 3 , 2020During task performance, brain states change dynamically and can appear recurrently. Recently, recurrent neural networks (RNN) have been used for identifying functional signatures underlying such brain states from task functional Magnetic Resonance Imaging (fMRI) data. While RNNs only model temporal dependence between time points, brain task decoding needs to model temporal dependencies of the underlying brain states. Furthermore, as only a subset of brain regions are involved in task performance, it is important to consider subsets of brain regions for brain decoding. To address these issues, we present a customised neural network architecture, Salient Patterns Over Time and Space (SPOTS), which not only captures dependencies of brain states at different time points but also pays attention to key brain regions associated with the task. On language and motor task data gathered in the Human Connectome Project, SPOTS improves brain state prediction by 17% to 40% as compared to the baseline RNN model. By spotting salient spatio-temporal patterns, SPOTS is able to infer brain states even on small time windows of fMRI data, which the present state-of-the-art methods struggle with. This allows for quick identification of abnormal task-fMRI scans, leading to possible future applications in task-fMRI data quality assurance and disease detection. Code is available at https://github.com/SCSE-Biomedical-Computing-Group/SPOTS.
@inproceedings{chan2020decoding, title = {Decoding task states by spotting salient patterns at time points and brain regions}, author = {Chan, Yi Hao and Gupta, Sukrit and Kasun, LL Chamara and Rajapakse, Jagath C}, booktitle = {Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology: Third International Workshop, MLCN 2020, and Second International Workshop, RNO-AI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4--8, 2020, Proceedings 3}, pages = {88--97}, year = {2020}, organization = {Springer}, }
2019
- Biomarkers
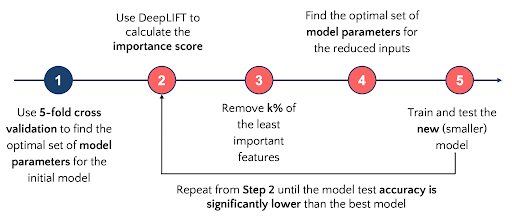 Decoding brain functional connectivity implicated in AD and MCISukrit Gupta, Yi Hao Chan, Jagath C Rajapakse , and 1 more authorIn International conference on medical image computing and computer-assisted intervention (Oral) , 2019
Decoding brain functional connectivity implicated in AD and MCISukrit Gupta, Yi Hao Chan, Jagath C Rajapakse , and 1 more authorIn International conference on medical image computing and computer-assisted intervention (Oral) , 2019Deep neural networks have been demonstrated to extract high level features from neuroimaging data when classifying brain states. Identifying salient features characterizing brain states further refines the focus of clinicians and allows design of better diagnostic systems. We demonstrate this while performing classification of resting-state functional magnetic resonance imaging (fMRI) scans of patients suffering from Alzheimer’s Disease (AD) and Mild Cognitive Impairment (MCI), and Cognitively Normal (CN) subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). We use a 5-layer feed-forward deep neural network (DNN) to derive relevance scores of input features and show that an empirically selected subset of features improves accuracy scores for patient classification. The common distinctive salient brain regions were in the uncus and medial temporal lobe which closely correspond with previous studies. The proposed methods have cross-modal applications with several neuropsychiatric disorders.
@inproceedings{gupta2019decoding, title = {Decoding brain functional connectivity implicated in AD and MCI}, author = {Gupta, Sukrit and Chan, Yi Hao and Rajapakse, Jagath C and Initiative, Alzheimer’s Disease Neuroimaging}, booktitle = {International conference on medical image computing and computer-assisted intervention}, pages = {781--789}, year = {2019}, organization = {Springer}, }