Evaluating XAI algorithms
Measuring robustness of explanations objectively
tl;dr: We propose RE-CONFIRM, a framework for evaluating explanations produced by XAI algorithms in an objective manner.
- RE-CONFIRM comprises 8 metrics that encompasses various aspects of a typical ML pipeline (data, models, labels).
- Using this framework on 2 fMRI datasets, we found that the combination of GAT and GNNExplainer produced explanations that are relatively more robust than other combinations considered in our study.
- Future biomarker discovery studies (that are based on applying explainers to deep learning models) can use RE-CONFIRM to determine the robustness of their model’s explanations.
Edit: Here’s is the link to our paper, which includes an additional set of experiments on top of those presented here. More details will be shared soon. :)
Problem
Numerous explainable AI algorithms have been developed and applied on biological datasets to derive potential biomarkers (i.e. salient features unique to a certain disorder/trait). While they can compute a score for each input feature to indicate its importance for the task (e.g. healthy subject vs patient with disorders), some studies have shown that these scores do not consistently produce explanations that are sensible for the task
Furthermore, there are few tools available to evaluate the robustness of these scores. Over the past years, model explainability has evolved into an essential component of recent research papers on using machine learning algorithms on biomedical datasets. While many papers are focused on creating deep / machine learning techniques that surpass the state-of-the-art, showing that the proposed model performs better (e.g. higher accuracy, lower error) is no longer sufficient - it is often expected for these studies to also highlight the salient features that the model identified to be important for performing the task.
Unfortunately, such analyses are often done at surface level, limited to highlighting the top 10 features, followed by rather limited cross-referencing with existing literature (e.g. mentioning that a subset of these top features were mentioned in some previous research paper, but often neglecting those that are not). One reason for this is the lack of standardised checklists or metrics for evaluating the robustness of the explanations produced by their proposed model
Existing solutions
Existing tools to evaluate the robustness of explainers typically came from research that highlighted issues with existing explainers. For example, Adebayo et al.
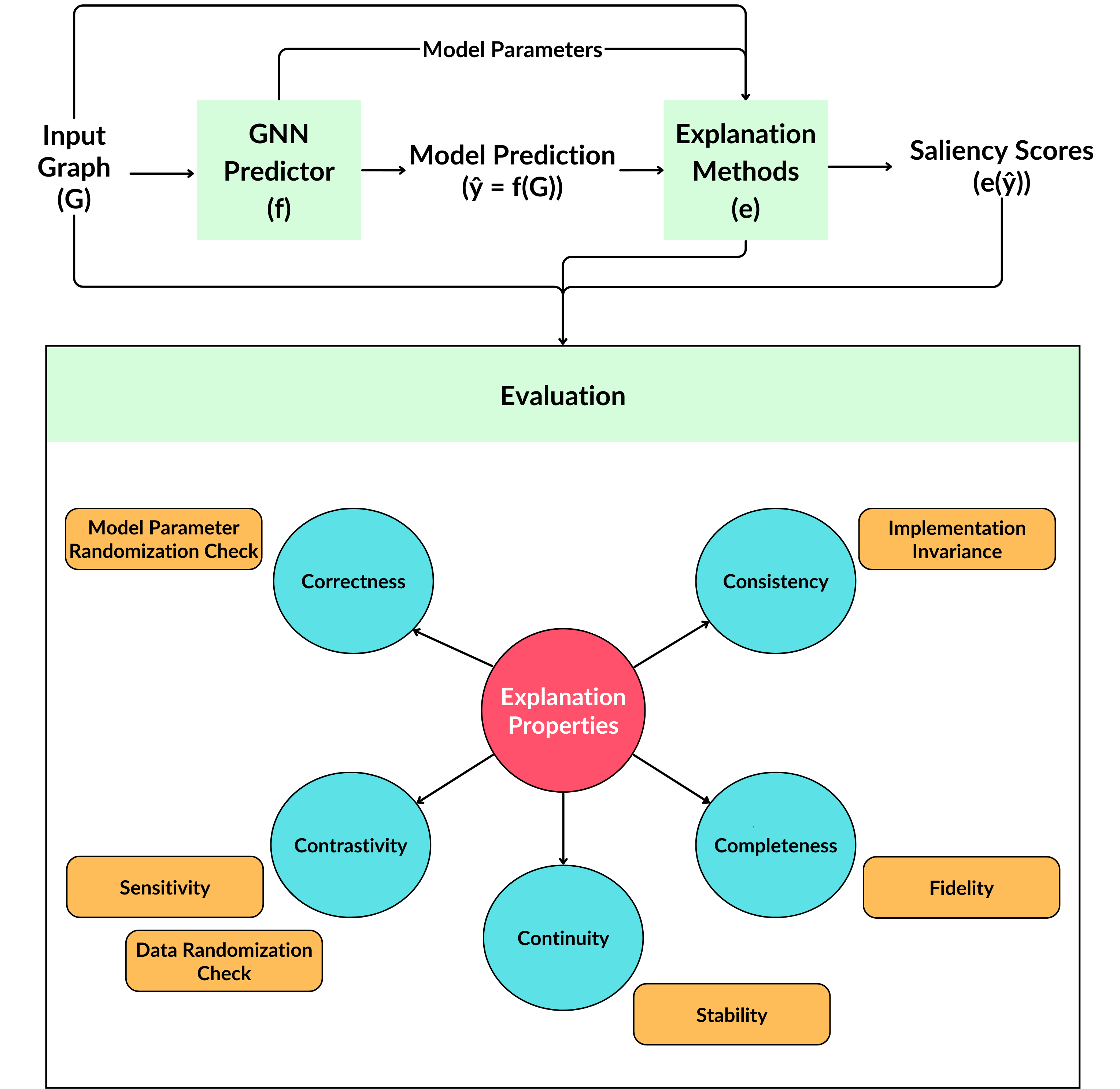
Since then, several tests have been developed that encompasses various aspects of a typical machine learning model (e.g. data, model weights, labels). These can be categorised into the following categories as proposed by Nauta et al.
- Correctness: Whether the explanation is faithful to the model ; e.g. Model Parameter Randomisation Check (MPRC) - if the explanation remains the same even when the model weights are randomised, then it is not considered to be faithfully correct.
- Consistency: Whether explainers are deterministic + models with same inputs and outputs should give the same explanation ; e.g. Implementation Invariance (II) - models with same architecture but different initialisation (seed) should have very similar explanations.
- Completeness: Whether the explanation captures the entire model behaviour ; e.g. Fidelity+ - retain a subset of the most important features, train another model on this subset and check whether the change in prediction is small.
- Continuity: Small changes in input that does not change the output much should not affect the explanation greatly too ; e.g. Stability measures the changes in model explanations when perturbations are introduced to the input.
- Contrastivity: Whether the explanation is discriminative with respect to targets; e.g. Data Randomisation Check (DRC) - a model trained on a dataset with shuffled labels should have very different explanations ; Sensitivity - explanations for each class should be distinct.
These categories are a subset of the full Co-12 framework they proposed. Other categories such as compactness, coherence and controllability are not considered as they are less relevant for biomarker discovery.
Methods
In this study, we attempt to make progress on developing a set of metrics that could serve as a standardised checklist for future research that applies ML to biological datasets.
We focus on two open source datasets: ABIDE and ADHD-200. These datasets contain over 700 functional magnetic resonance imaging (fMRI) scans along with phenotypic data (demographics, clinical test scores, presence/absence of disorders). Functional connectivity matrices are often derived from them and used as inputs to machine learning models.
We use a subset of Nauta’s Co-12 framework as discussed above. These existing evaluation metrics are rather generic. Thus, we additionally proposed 2 novel metrics that capture topological properties characteristic of brain functional connectomes. More information about them will be revealed when the paper is published.
Put together, these 8 metrics form the basis of our proposed RE-CONFIRM framework.
In our implementation of the metrics, we measured the changes in explanations quantitatively using Hellinger Distance (for MPRC, DRC, Stability, Sens, II). For Fidelity, we focused on the top 20 features. To evaluate stability, we perturbed the input graph data by rewiring edges between second-degree nodes
Results
We experimented with 3 predictors and 4 explainers, namely:
Predictors: (i) ChebGCN, (ii) GraphSAGE, (iii) Graph Attention Network (GAT). 2 hidden layers were used for all models.
Explainers: (i) Integrated Gradients (IG), (ii) Guided Backpropagation (GBP), (iii) GNNExplainer (GNNExp), (iv) Attention.
Saliency scores can be influenced by a multitude of factors: (i) the base model, (ii) the explainer and (iii) the dataset. Thus, 3 sets of experiments were performed to evaluate (i) Performance across predictors, (ii) Performance across explainers, (iii) Performance across datasets.
Click here to view the result tables
| ABIDE | ADHD | |||||||
|---|---|---|---|---|---|---|---|---|
| Metrics | GAT | SAGE | GCN | Cheb | GAT | SAGE | GCN | Cheb |
| MPRC (↑) | 0.488 | 0.372 | 0.162 | 0.379 | 0.300 | 0.248 | 0.411 | 0.207 |
| DRC (↑) | 0.235 | 0.123 | 0.177 | 0.152 | 0.241 | 0.162 | 0.156 | 0.137 |
| Fidelity+ (↑) | 0.330 | 0.044 | 0.023 | 0.092 | 0.292 | 0.202 | 0.349 | 0.237 |
| Stability (↑) | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Fidelity- ( ↓ ) | 0.101 | 0.008 | 0.006 | 0.031 | 0.113 | 0.075 | 0.102 | 0.099 |
| Sens ( ↓ ) | 0.011 | 0.008 | 0.010 | 0.008 | 0.009 | 0.011 | 0.011 | 0.004 |
| II ( ↓ ) | 0.009 | 0.030 | 0.022 | 0.029 | 0.015 | 0.021 | 0.011 | 0.021 |
Table 1. Evaluation metrics for GNNExplainer on both datasets (whole).
| GNNExp | GBP | IG | Attn | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metric | GAT | GCN | GAT | SAGE | GCN | Cheb | GAT | SAGE | GCN | Cheb | GAT |
| MPRC (↑) | 0.300 | 0.411 | 0.316 | 0.137 | 0.278 | 0.137 | 0.086 | 0.144 | 0.365 | 0.316 | 0.166 |
| DRC (↑) | 0.241 | 0.156 | 0.270 | 0.132 | 0.174 | 0.118 | 0.179 | 0.196 | 0.194 | 0.605 | 0.160 |
| Fidelity+ (↑) | 0.292 | 0.349 | 0.197 | 0.135 | 0.152 | 0.168 | 0.287 | 0.181 | 0.212 | 0.179 | 0.127 |
| Stability (↑) | 1.000 | 1.000 | 0.999 | 1.000 | 1.000 | 1.000 | 0.452 | 0.697 | 0.728 | 0.793 | 0.847 |
| Fidelity- (↓) | 0.113 | 0.102 | 0.133 | 0.104 | 0.122 | 0.142 | 0.248 | 0.200 | 0.178 | 0.150 | 0.117 |
| Sens (↓) | 0.009 | 0.011 | 0.008 | 0.009 | 0.008 | 0.007 | 0.038 | 0.022 | 0.024 | 0.009 | 0.084 |
| II (↓) | 0.015 | 0.011 | 0.015 | 0.010 | 0.008 | 0.009 | 0.016 | 0.007 | 0.017 | 0.013 | 0.138 |
Table 2. Evaluation metrics for GNNExplainer, GBP and IG on ADHD-200 (whole).
| ABIDE | ADHD | |||||||
|---|---|---|---|---|---|---|---|---|
| Metric | GAT | SAGE | GCN | Cheb | GAT | SAGE | GCN | Cheb |
| MPRC (↑) | 0.322 | 0.564 | 0.169 | 0.144 | 0.282 | 0.319 | 0.423 | 0.166 |
| DRC (↑) | 0.233 | 0.160 | 0.197 | 0.153 | 0.193 | 0.086 | 0.168 | 0.168 |
| Fidelity+ (↑) | 0.250 | 0.260 | 0.037 | 0.228 | 0.413 | 0.073 | 0.300 | 0.035 |
| Stability (↑) | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Fidelity- (↓) | 0.052 | 0.054 | 0.011 | 0.069 | 0.109 | 0.035 | 0.097 | 0.011 |
| Sens (↓) | 0.023 | 0.009 | 0.011 | 0.012 | 0.010 | 0.006 | 0.015 | 0.011 |
| II (↓) | 0.012 | 0.009 | 0.017 | 0.013 | 0.011 | 0.011 | 0.012 | 0.009 |
Table 3. Metrics for GNNExplainer on both datasets (site NYU only).
Performance across predictors: We found that GAT performs the best, with much higher MPRC, DRC and Fidelity+ than other models. GCN was found to be relatively erratic as it has the highest MPRC and Fidelity+ for ADHD, but lowest in ABIDE. Overall, positive findings for all models include low Fidelity-, very high stability and low II, However, all models have low DRC, Fidelity+ and target sensitivity.
Performance across explainers: GNNExplainer generally performs the best across most metrics and this finding is also replicated in ABIDE. Attention explainer and IG have much lower stability and MR than others, while GBP has lower MPRC. Notably, the previous finding of low Fidelity- and II remains, but all explainers still have low DRC, Fidelity+ (except GNNExplainer) and target sensitivity.
Performance across datasets: Comparing the results from the whole dataset, it is evident that the metrics are sensitive to dataset size
Discussion
Key findings of our experiments include:
- GNNExplainer and GAT exhibits greatest robustness amongst all combinations.
- Low Fidelity- and II across all combinations suggests that unimportant features are correctly given low scores and the scores are consistent.
- Low target sensitivity and DRC indicates poor contrastivity, raising concerns about whether the saliency features identified for the disease class are actually unique to them (as expected from a strong biomarker)
- GBP was found to have generally low MPRC, which aligns with findings in Adebayo et al. that GBP is invariant to their randomisation tests in computer vision datasets.
- IG and attention explainer have consistently lower stability, suggesting that they might not be a good choice for biomarker discovery.
Future work in this research direction could consider that:
- Our study is limited to explaining the model’s decision ; alternatively, one could further study the ‘phenomenon’ (i.e. use the ground truth labels as the target instead of the model’s predictions)
to corroborate these findings. - Our study is focused on post-hoc gradient-based explainers
Deep neural networks are often seen as black boxes - most of them do not provide an intuitive understanding of how they arrive at a decision (in contrast to decision trees, where the criteria used to split the dataset is clearly represented in each node). Post-hoc explainers are limited to highlighting input-output relationships and do not give insight to the models beyond that (unlike decision trees). However, for the purpose of biomarker discovery, identifying the most salient features would be sufficient for the task. . Perturbation-based approaches and other intrinsically interpretable models (e.g. graph pooling) could be studied in future.
Code
Our code repository will be released soon!
We hope that RE-CONFIRM will serve as the first step towards a standard for quantitative evaluation of salient features detected by future studies.